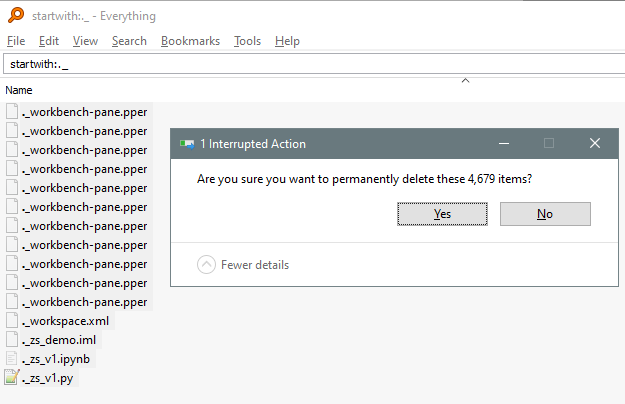
EERC is counterpoint_id 606. Could select by bounding box (lat/long) too. This includes a list of counts (just labeled “id”) but no actual counts, so you could do time/date collection stat comparisons from there as well.
It looks like minutes start/end on the second that the count was started, so aggregation by minute (not second) should be used.
It looks like minutes without data are excluded, e.g. no 12:51:52 minute is included here (EERC 606):
{"id":281510,"count_session_id":18223,"timestamp":"2016-06-21T12:51:56.000Z","bike_generic":0,"bike_adult":0,"bike_child":0,"bike_baby":0,"bike_cargo":0,"bike_child_trailer":0,"walk_generic":0,"walk_adult":0,"walk_child":0,"walk_stroller":0,"walk_disability":0,"walk_skateboard":0,"walk_visually_impaired":0,"walk_physically_impaired":0,"vehicle_personal":2,"vehicle_car":0,"vehicle_semi":0,"vehicle_moto":0,"vehicle_transit":0,"vehicle_oversized":0,"objectId":null,"gender_m":0,"gender_f":0,"gender_o":0,"onePerson":0,"twoPerson":0,"threePerson":0,"fourPlusPerson":0},{"id":281511,"count_session_id":18223,"timestamp":"2016-06-21T12:53:56.000Z","bike_generic":0,"bike_adult":0,"bike_child":0,"bike_baby":0,"bike_cargo":0,"bike_child_trailer":0,"walk_generic":0,"walk_adult":0,"walk_child":0,"walk_stroller":0,"walk_disability":0,"walk_skateboard":0,"walk_visually_impaired":0,"walk_physically_impaired":0,"vehicle_personal":5,"vehicle_car":0,"vehicle_semi":0,"vehicle_moto":0,"vehicle_transit":0,"vehicle_oversized":0,"objectId":null,"gender_m":0,"gender_f":0,"gender_o":0,"onePerson":0,"twoPerson":0,"threePerson":0,"fourPlusPerson":0}
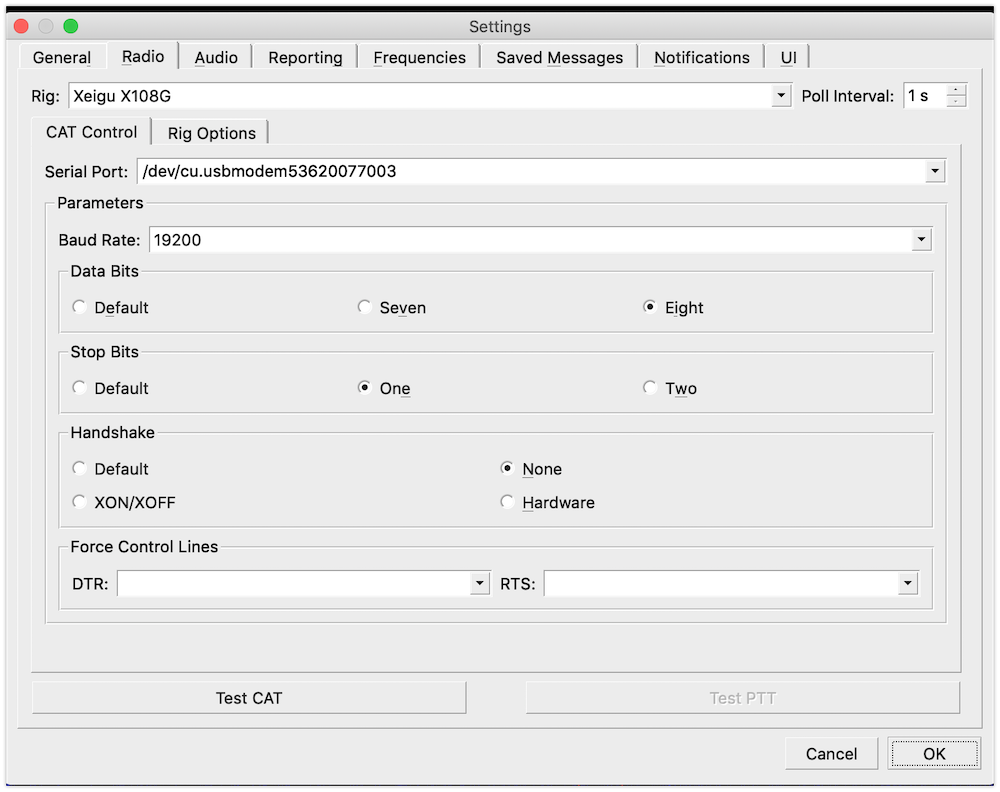 Following this post by HB9TXB Branko, I did also check the list of serial ports using this command in the terminal: `ls -lrt /dev/tty*`.
Following this post by HB9TXB Branko, I did also check the list of serial ports using this command in the terminal: `ls -lrt /dev/tty*`.